Image data science with Python and Napari#
A first Latin America workshop #LIBREhub @napari #LABI - Latin America Bioimaging #MexicoBioimaging
This Jupyter book contains training resources for scientists who want to dive into image processing with Python and Napari. It specifically aims for scientists and students working with microscopy images in the life sciences. We presume the attendees have some basic python programming and image analysis knowledge. To get everyone on the same level, we start with Python programming basics. We will process images using numpy, scipy, scikit-image, SimpleITK and clEsperanto. We will explore Napari for interactive image data analysis and the Napari-Assistant for generating Jupyter Notebooks from interactively designed image processing workflows.
Note: Before the course starts, all partcipants are required to have Napari and Jupyter installed, please see the course preparation page - and make use of the napari chatroom for troubleshooting if you face installation issues. The session on Python environments is further more in the inverted classroom format, so we kindly ask that you familiarise yourself with the dedicated teaching material on environments before the session in order to simply use the given time for questions.
Timetable#
All following times are in Chilean (Santiago) time (currently GMT-4). Look up your related time here, so you don’t miss anything.
Day 1 - Monday the 7th of August 2023#
Time (Chile) |
Activities / Topics |
Presenters |
Support |
|---|---|---|---|
08.00 – 09.00 |
Motivating talk - Napari bioimage analysis |
– |
|
09.00 – 09.55 |
Welcome and ice-break Session |
Pierre Padilla Huamantico |
|
10.10 – 12.00 |
Environments Q & A |
Marcelo Zoccoler, |
|
12:00 - 13:00 |
Lunch Break |
||
13.00 – 14.55 |
Introduction to the napari assistant |
Marcelo Zoccoler |
|
15.10 – 16.30 |
• Pitfalls when working with Jupyter notebooks |
Adan Guerrero Cardenas, |
|
16.30 – 17.30 |
• Masking numpy arrays |
Adan Guerrero Cardenas, |
Day 2 - Wednesday the 9th of August 2023#
Time (Chile) |
Activities / Topics |
Presenters |
Support |
|---|---|---|---|
10:30 - 11:00 |
Questions from the last day? |
Tobias Wenzel, |
|
11.00 – 12.00 |
Python advanced folder management with loops |
Adan Guerrero Cardenas, |
|
12.00 – 14.00 |
Tips for open source image analysis |
– |
|
14:00 - 16:00 |
Lunch Break |
||
16.00 – 19.00 |
Image analysis under the hood with python and napari: |
Adan Guerrero Cardenas, |
If you run the exercises in your own time and questions arise, please ask on https://forum.image.sc/.
Day 3 - Friday the 11th of August 2023 open to all here on Zoom#
On the third and last day, there are showcase seminars of key napari plugins as well as emerging trends to controll your bioimaging hardware with python libraries.
Time |
Activities / Topics |
Presenters |
|---|---|---|
07.55 – 08.00 |
Welcome! |
|
08.00 – 08.20 |
Napari Animation |
|
08:20-8:50 |
Affinder |
|
08:50 – 9:30 |
Napari Clusters Plotter (Unsupervised machine learning) |
|
09:30 - 10:00 |
Break |
|
10.10 – 11.00 |
Napari Zelda (3D segmentation) |
|
11.00 – 12.00 |
Napari Superres (Superresolution) |
|
12:00 - 12:40 |
Napari real time processing |
|
12:40 - 14:00 |
Lunch Break |
|
14:00 - 15:00 |
Python libraries for microscope control |
How to use this material#
For following the course, we recommend downloading the repository from which this Jupyter book is made. All Jupyter Notebooks are executable so that attendees can reproduce all demos and exercises.
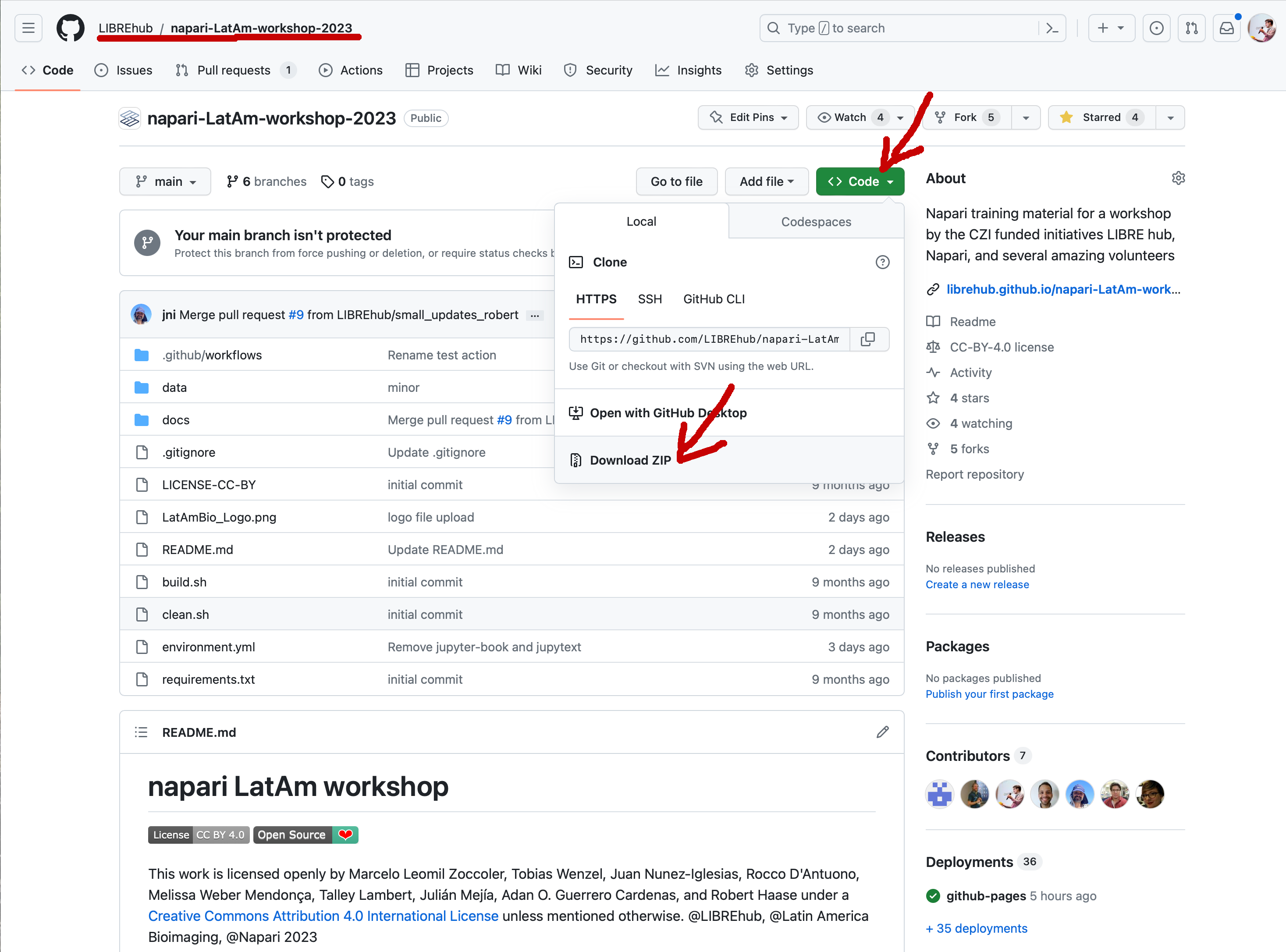
Assuming you downloaded the repository to your Desktop, you can open the Jupyter book by opening a terminal and typing:
cd Desktop/napari-LatAm-workshop-2023
micromamba activate napari-latam
jupyter lab
If you do not yet have conda or devbio-napari-env installed, first follow the “Course preparation” installation instructions on the next page.
Using Jupyter lab, you can navigate to the course lessons in the docs folder.
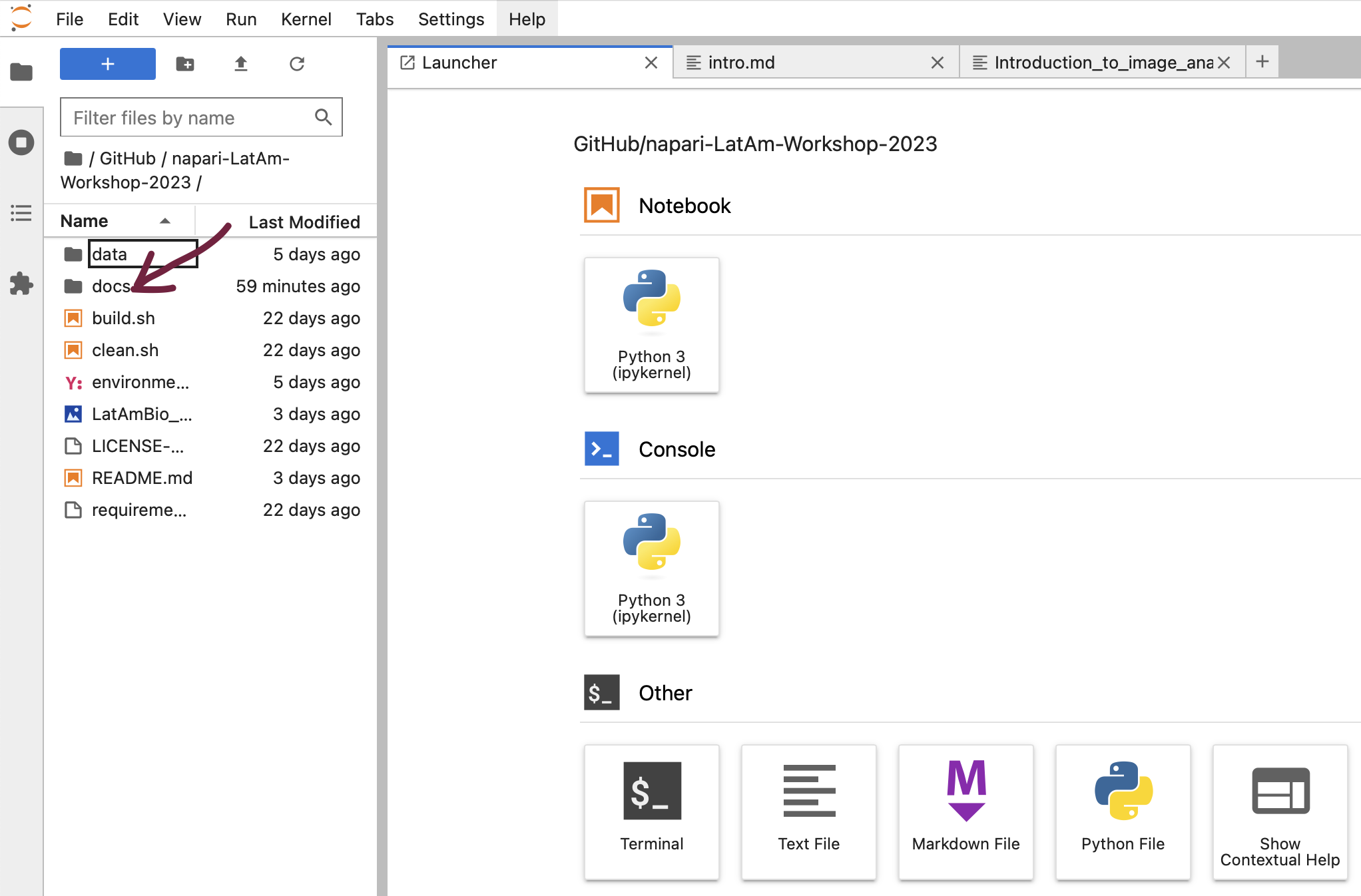
… and execute the code and experiment with it.
Feedback and support#
If you have any questions, please create a github issue. Alternatively, open a thread on image.sc.
Acknowledgements#
This course was held virtually at the Institute for Biological and Medical Engineering, Universidad Católica de Chile in August 2023 as part of the LIBRE hub project. We would like to thank all online contributors and speakers for their support and the Chan Zuckerberg Initiative for financial support through the LIBRE hub project. We would like to thank all the people who shared teaching materials we are reusing here, in particular from an EPFL copurse last year and individual contributors including Marcelo Leomil Zoccoler, Tobias Wenzel, Juan Nunez-Iglesias, Rocco D’Antuono, Melissa Weber Mendonça, Talley Lambert, Julián Mejía, Adan O. Guerrero Cardenas, and Robert Haase.
